Data Science Bowl 2018 Recap
18 May 2018 This year our team competed in the 2018 Kaggle Data Science Bowl. The goal of
the competition was to identify cells in microscopic images. There can be zero,
one or many cells in any given image. Our solution was a Convolutional Neural
Network with skip connections.
This year our team competed in the 2018 Kaggle Data Science Bowl. The goal of
the competition was to identify cells in microscopic images. There can be zero,
one or many cells in any given image. Our solution was a Convolutional Neural
Network with skip connections.
We used Google Colab which is a free Jupyter notebook-like environment with a GPU. It offers realtime collaboration similar to google docs and google sheets. The Overview of Colaboratory Features is a good resource to learn this tool as well as the example notebooks.
The dataset provided consisted of png images and masks.
stage1_train
├── 00071198d059ba7f5914a526d124d28e6d010c92466da21d4a04cd5413362552
│ ├── images
│ │ └── 00071198d059ba7f5914a526d124d28e6d010c92466da21d4a04cd5413362552.png
│ └── masks
│ ├── f6eee5c69f54807923de1ceb1097fc3aa902a6b20d846f111e806988a4269ed0.png
│ ├── ffae764df84788e8047c0942f55676c9663209f65da943814c6b3aca78d8e7f7.png
│ ...
├── 003cee89357d9fe13516167fd67b609a164651b21934585648c740d2c3d86dc1
│ ├── images
│ │ └── 003cee89357d9fe13516167fd67b609a164651b21934585648c740d2c3d86dc1.png
│ └── masks
│ ├── 23947be0224e633947e9a3935d8ecf89be3eb56f68f30db85c74d4bf07d3028b.png
│ ├── 2a709842ce867d9b27281f9d8bec4968b88805afdff3de2ed0c8156f711483cd.png
│ ...
...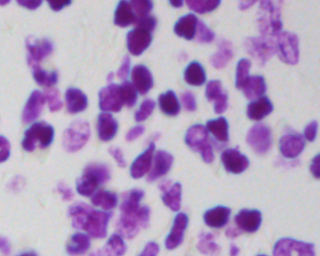 Each training example would be provided as a directory containing the image as
captured from the device.
Each training example would be provided as a directory containing the image as
captured from the device.
 And a set of masks showing where a cell was identified by an expert. For every
training example you would be provided between 1 and ~300 masks.
And a set of masks showing where a cell was identified by an expert. For every
training example you would be provided between 1 and ~300 masks.
The submission criteria required us to Run Length Encode the model’s predictions. It is a technique that can communicate large runs of information more efficiently than jpeg.
The approach we used was the same one as the winner of the ISBI cell tracking challenge 2015: The U-net architecture. It uses skip connections similar to LSTMs to keep track of ‘context’ in 2 dimensions. This context improves the performance of the model because it can better reason about the textures it is currently looking at while considering if it is looking at the inside of a cell or looking at a non-cell region of space.
To learn more about skip connections in NNs see Deep Learning Section 10.9.1 and Understanding LSTM Networks.
The U-net paper mentions Upsampling. This is the same as the term “Deconvolution” used in other Deep Learning literature. Deconvolution can be done in many ways. The method that ships with Keras is the Conv2DTranspose.
Throughout the competition we learned the loss function is critical to the performance of the network. It may sound obvious but careful selection of an appropriate loss function will change the way the model behaves. For example, we engineered the loss function to segment cells better by giving larger penalty for misclassifying the cell boundaries. This worked because many cell images have cells overlapping and touching. Early versions of the model would have trouble learning the boundaries between cells. These boundaries should have been labeled as not being a cell, but would incorrectly be classified as a cell.
We experimented with removing padding. The accuracy of the model went down. We believe this is because traditionally padding is used make the model give consideration to the pixels near the border of the image. This means that the border of the images have information that is useful for prediction.
Jacob compiled separate notes on Convolutional NNs.
Our work is visible at N.Y.C. Kaggle DS Bowl Colab, github and our google drive.
Congratulations to this years winning kernel.
And thank you to teammates Taraqur Rahman, Jacob Peters and Cal Almodovar.
If you need help solving your business problems with software read how to hire me.
comments powered by Disqus